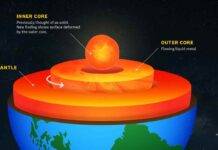
With Mars and Europa out of reach, many scientists have turned to studying some of the Arctic and Antarctic microbes that have adapted to similarly harsh conditions on Earth.
One recent study has traced the evolutionary branches of Arctic bacterial resistance to toxic mercury—an adaptation that appears to have an ancient lineage. The results of a previous expedition to the Arctic found that up to 31 percent of bacteria retrieved from various locations and grown in lab cultures contain the mercuric reductase gene(merA), a genetic sequence that encodes an enzyme that is capable of breaking down toxic mercury into a more harmless chemical form. That’s a crucial survival trait, as growing mercury emissions from human sources add to natural sources to dump more than 300 tons of the toxic contaminant in the Arctic every year. The latest research finds evidence of merA having both recent and ancient evolutionary lineages among the samples of Arctic bacteria.
“This suggests that merA has been present in the High Arctic for an extended time period, and that mercury contamination of the Arctic is not a new phenomenon,” said Niels Kroer, a microbiologist and head of the Department of Environmental Science at Aarhus University in Denmark. “In other words, transport of mercury to the high Arctic by the atmosphere is a natural process predating the Industrial Revolution.”
The merA resistance works by reducing the Hg(II) form of mercury to Hg(0). The latter represents an elemental form of mercury that can evaporate into the atmosphere and lead to detoxification of the bacteria’s immediate environment. Chemical reduction through exposure to sunlight does most of the mercury removal work in snow, but Kroer’s team of Danish and U.S. researchers previously estimated that the mercury-resistant bacteria help remove between 2 and 10 percent of the mercury in Arctic snow. Their recent work, detailed in the journal FEMS Microbiology Ecology, looked more closely at the diversity of merA genes among mercury-resistant bacteria in the Arctic.

The researchers may not have had to travel to Mars, but they still faced a tough journey to the sampling site at Station Nord, located about 575 miles south of the North Pole in Greenland. Kroer’s first attempt to fly to the site aboard a C-130 Hercules military aircraft of the Danish Air Force in early April failed when bad weather prevented it from landing. A second successful attempt in May coincided with good weather—sunshine 24 hours a day and a brisk temperature of -25 degrees Celsius—that allowed the researchers to gather samples from high Arctic snow, freshwater and sea-ice brine.
Genetic testing found seven different varieties of merA genes among the 71 mercury-resistant bacterial samples, including three new varieties of previously undiscovered merA genes. But the biggest surprise of the study came from finding merA genes among just 5 of the 18 bacteria samples that showed they could reduce Hg(II) to Hg(0). The finding suggests there are likely many more undiscovered merA genes in the Arctic beyond the three newly identified varieties.
The discovery of the same merA gene variants among many different taxonomic subgroups of bacteria indicates that many Arctic microbes gained their mercury resistance through horizontal transfer—the swapping of DNA molecules, called plasmids, which can self-replicate independently of the main chromosomes. Known merA varieties can be found around the world at locations as diverse as sugar beet leaves in the UK, to 120,000-year-old Siberian permafrost samples, and within a mercury mine in Central Asia.
The spread of merA genes among Arctic bacteria likely reflects a combination of such horizontal transfers, local selective pressure, and certain common DNA sequences that have been conserved across species despite millions of years of evolution, Kroer said. A whole-genome sequencing test of the bacterium Flavobacterium SOK62 also revealed the presence of an arsR regulator gene often associated with more ancient mercury-resistant lineages.
Kroer’s group also found a wide range of mercury resistance within different Arctic sub-environments. Less than 2 percent of the sample bacteria from an ice-covered freshwater lake and sea-ice brine demonstrated mercury resistance. By comparison, almost one-third of the snow bacteria samples were resistant to mercury.
The latest study is just a small step toward building a more comprehensive picture of the distribution of merA genes among Arctic microbes. Kroer anticipates more studies would need to be done at a variety of other locations, as well as more whole-genome sequencing tests that could help identify more mystery merA varieties among Arctic microbes. That knowledge could also help researchers better understand the processes leading to the buildup of mercury in life forms such as seals and polar bears.
In a broader sense, understanding the evolutionary lineage of genes that help organisms adapt to harsh environments has bigger-picture implications for understanding life’s survival in the broader universe. It’s no accident that Kroer’s recent research on Arctic microbes was partly funded by the NASA Exobiology and Evolutionary Biology Program.
“Various physiological adaptations, such as increased membrane fluidity, synthesis of cold- adapted enzymes and production of cold shock and antifreeze proteins enable bacteria to survive under cold conditions, and bacterial activity has been detected at sub-zero temperatures in sea ice and snow,” Kroer said. “The same mechanism may also be important for extraterrestrial life.”
More information:
Møller, A. K., Barkay, T., Hansen, M. A., Norman, A., Hansen, L. H., Sørensen, S. J., Boyd, E. S. and Kroer, N. (2014), “Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine.” FEMS Microbiology Ecology, 87: 52–63. doi: 10.1111/1574-6941.12189
Note : The above story is based on materials provided by Astrobio.net