Paleontologists have documented the evolutionary adaptations necessary for ancient lobe-finned fish to transform pectoral fins used underwater into strong, bony structures, such as those of Tiktaalik roseae. This enabled these emerging tetrapods, animals with limbs, to crawl in shallow water or on land. But evolutionary biologists have wondered why the modern structure called the autopod–comprising wrists and fingers or ankles and toes–has no obvious morphological counterpart in the fins of living fishes.
In the Dec. 22, 2014, issue of the Proceedings of the National Academy of Sciences, researchers argue previous efforts to connect fin and fingers fell short because they focused on the wrong fish. Instead, they found the rudimentary genetic machinery for mammalian autopod assembly in a non-model fish, the spotted gar, whose genome was recently sequenced.
“Fossils show that the wrist and digits clearly have an aquatic origin,” said Neil Shubin, PhD, the Robert R. Bensley Professor of organismal biology and anatomy at the University of Chicago and a leader of the team that discovered Tiktaalik in 2004. “But fins and limbs have different purposes. They have evolved in different directions since they diverged. We wanted to explore, and better understand, their connections by adding genetic and molecular data to what we already know from the fossil record.”
Initial attempts to confirm the link based on shape comparisons of fin and limb bones were unsuccessful. The autopod differs from most fins. The wrist is composed of a series of small nodular bones, followed by longer thin bones that make up the digits. The bones of living fish fins look much different, with a set of longer bones ending in small circular bones called radials.
The primary genes that shape the bones, known as the HoxD and HoxA clusters, also differ. The researchers first tested the ability of genetic “switches” that control HoxD and HoxA genes from teleosts–bony, ray-finned fish–to shape the limbs of developing transgenic mice. The fish control switches, however, did not trigger any activity in the autopod.
Teleost fish–a vast group that includes almost all of the world’s important sport and commercial fish–are widely studied. But the researchers began to realize they were not the ideal comparison for studies of how ancient genes were regulated. When they searched for wrist and digit-building genetic switches, they found “a lack of sequence conservation” in teleost species.
They traced the problem to a radical change in the genetics of teleost fish. More than 300 million years ago, after the fish-like creatures that would become tetrapods split off from other bony fish, a common ancestor of the teleost lineage went through a whole-genome duplication (WGD)–a phenomenon that has occurred multiple times in evolution.
By doubling the entire genetic repertoire of teleost fish, this WGD provided them with enormous diversification potential. This may have helped teleosts to adapt, over time, to a variety of environments worldwide. In the process, “the genetic switches that control autopod-building genes were able to drift and shuffle, allowing them to change some of their function, as well as making them harder to identify in comparisons to other animals, such as mice,” said Andrew Gehrke, a graduate student in the Shubin lab and lead author of the study.
Not all bony fishes went through the whole genome duplication, however. The spotted gar, a primitive freshwater fish native to North America, split off from teleost fishes before the WGD.
When the research team compared Hox gene switches from the spotted gar with tetrapods, they found “an unprecedented and previously undescribed level of deep conservation of the vertebrate autopod regulatory apparatus.” This suggests, they note, a high degree of similarity between “distal radials of bony fish and the autopod of tetrapods.”
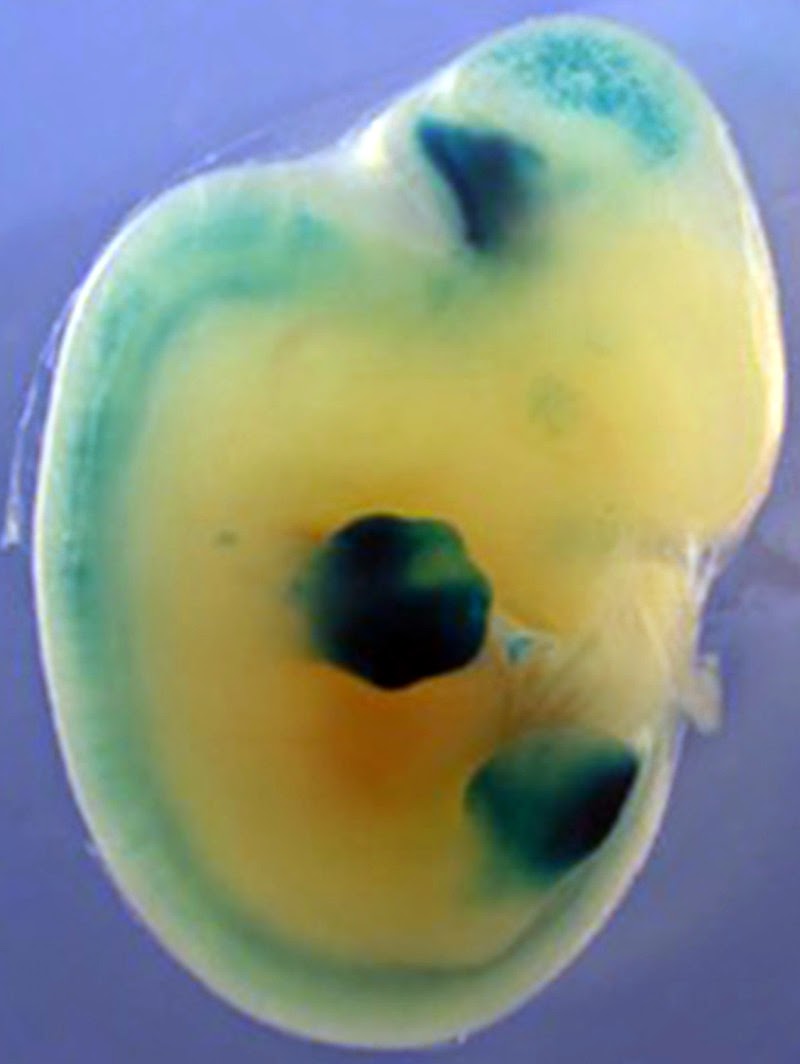
They tested this by inserting gar gene switches related to fin development into developing mice. This evoked patterns of activity that were “nearly indistinguishable,” the authors note, from those driven by the mouse genome.
“Overall,” the researchers conclude, “our results provide regulatory support for an ancient origin of the ‘late’ phase of Hox expression that is responsible for building the autopod.”
This study was supported by the Brinson Foundation; the National Science Foundation; the Brazilian National Council for Scientific and Technological Development grants; the National Institutes of Health; the Volkswagen Foundation, Germany; the Alexander von Humboldt-Foundation, the Spanish and Andalusian governments; and Proyecto de Excelencia.
Additional authors include Mayuri Chandran and Tetsuya Nakamura from the University of Chicago; Igor Schneider from the Instituto de Ciencias Biologicas, Universida de Federal do Para, Belem, Brazil; Elisa de la Calle-Mustienes, Juan J. Tena, Carlos Gomez-Marin and José Luis Gómez-Skarmeta from the Centro Andaluz de Biología del Desarrollo, Sevilla, Spain; and Ingo Braasch and John H. Postlethwait from the Institute of Neuroscience, University of Oregon.
Reference:
Andrew R. Gehrke, Igor Schneider, Elisa de la Calle-Mustienes, Juan J. Tena, Carlos Gomez-Marin, Mayuri Chandran, Tetsuya Nakamura, Ingo Braasch, John H. Postlethwait, José Luis Gómez-Skarmeta, and Neil H. Shubin. Deep conservation of wrist and digit enhancers in fish. PNAS, December 22, 2014 DOI: 10.1073/pnas.1420208112
Note : The above story is based on materials provided by University of Chicago Medical Center.










