A group scientists have used new genetic sequencing data to understand how an ancient organism that lived alongside the dinosaurs has evolved over millions of years. A four-year effort by a genetic research team from a dozen universities has uncovered for the first time the biology and evolution of dinoflagellates, tiny but complex organisms primarily known as marine plankton.
You can find dinoflagellates everywhere. They turn sunlight into oxygen for the air we breathe. They are the ancient building blocks of fossil fuel. They are found in dietary supplements in the grocery store to boost brain health. They make the lagoons glow at night in places like Puerto Rico. They are also responsible for toxic algal blooms that can kill off massive amounts of fish and toxins in shellfish that can harm humans.
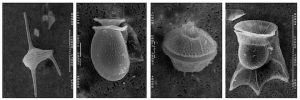
Even though they are a key species in the environment, they have long been misunderstood due to their large, complex genomes. A single dinoflagellate may have 12 to 400 chromosomes in its large nucleus. For comparison, humans have 23 pairs, totaling 46. The scientists were able to combine molecular, fossil, and biogeochemical evidence to map major landmarks in the evolution of dinoflagellates over millions of years, resulting in a revised model for their evolution.
“Mapping the evolution of dinoflagellates has never been done — successfully and on such a scale — before,” said Tsvetan Bachvaroff, a molecular geneticist at the University of Maryland Center for Environmental Science. He provided samples to the group of two kinds of parasitic dinoflagellates that he grows at the Institute of Marine and Environmental Technology in Baltimore.
“Now we understand how they are related, what they look like,” he said “It’s their genomic flexibility that has given them the advantage to evolve.”
“This work provides important new insights into the evolution of dinoflagellates and we are delighted that Dr. Bachvaroff was able to contribute significantly to this study,” said Russell Hill, director of the Institute of Marine and Environmental Technology.
The study found that nonphotosynthetic dinoflagellates, those that don’t use sunlight to synthesize nutrients from carbon dioxide and water, have retained plastids (the manufacturing site of important chemical compounds in the cells of plants and algae) that have vital metabolic functions, one of which may be the evolutionary source of dinoflagellate luminescence.
“The prediction that a big chunk of dinoflagellate diversity including all free-living heterotrophs have retained remnant plastids illustrates how little known they are and could help us better understand their roles in the oceans and other areas,” said the study’s lead author, Jan Janouskovec of the University College of London.
The findings could lead to a better understanding of how bioluminescence works, how to turn off harmful red tides, or how to identify areas rich with oil by looking at fossilized dinoflagellates in the rock.
“They violate the fundamental rules of Darwinian evolution. They tend to borrow genes from different places,” Bachvaroff said. “The ocean is like this giant parts catalog where from which you can grab genes and use them. Organisms that can acquire genes from their environment have a selective advantage.”
Reference:
Jan Janouškovec, Gregory S. Gavelis, Fabien Burki, Donna Dinh, Tsvetan R. Bachvaroff, Sebastian G. Gornik, Kelley J. Bright, Behzad Imanian, Suzanne L. Strom, Charles F. Delwiche, Ross F. Waller, Robert A. Fensome, Brian S. Leander, Forest L. Rohwer, Juan F. Saldarriaga. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proceedings of the National Academy of Sciences, 2017; 114 (2): E171 DOI: 10.1073/pnas.1614842114
Note: The above post is reprinted from materials provided by University of Maryland Center for Environmental Science.










